Quality Control
How to exclude low quality cells.
During dataset creation
The first opportunity for QC is when adding a new dataset:
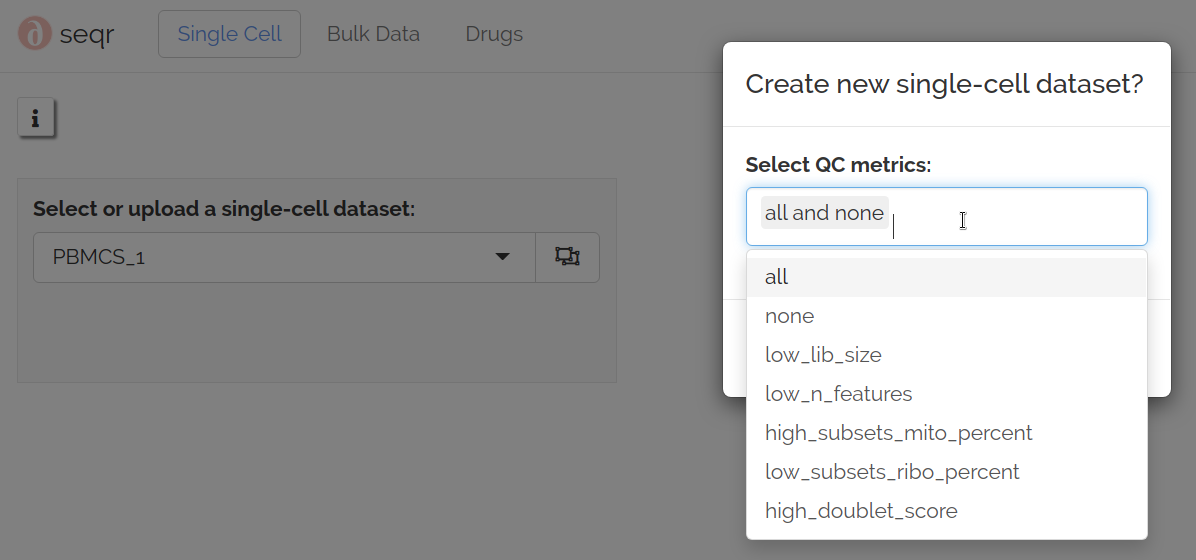
Metric descriptions:
- none: only remove empty droplets
- all (default): use all metrics
- all_and_none: both of above - creates datasets QC0 and QC1
- low_lib_size: remove cells that have low total counts
- low_n_features: remove cells that have few genes with counts
- high_mito_percent: remove cells with high mitochondrial percent
- low_ribo_percent: remove cells with low ribosomal percent
- high_doublet_score: remove cells called as doublets by scDblFinder.
The above metrics are calculated using outlier detection and applied prior to normalization.
- none: only remove empty droplets
- all (default): use all metrics
- all_and_none: both of above - creates datasets QC0 and QC1
- low_lib_size: remove cells that have low total counts
- low_n_features: remove cells that have few genes with counts
- high_mito_percent: remove cells with high mitochondrial percent
- low_ribo_percent: remove cells with low ribosomal percent
- high_doublet_score: remove cells called as doublets by scDblFinder.
The above metrics are calculated using outlier detection and applied prior to normalization.
After dataset creation
For an existing dataset, QC Score and Boolean Features can be plotted just like genes:
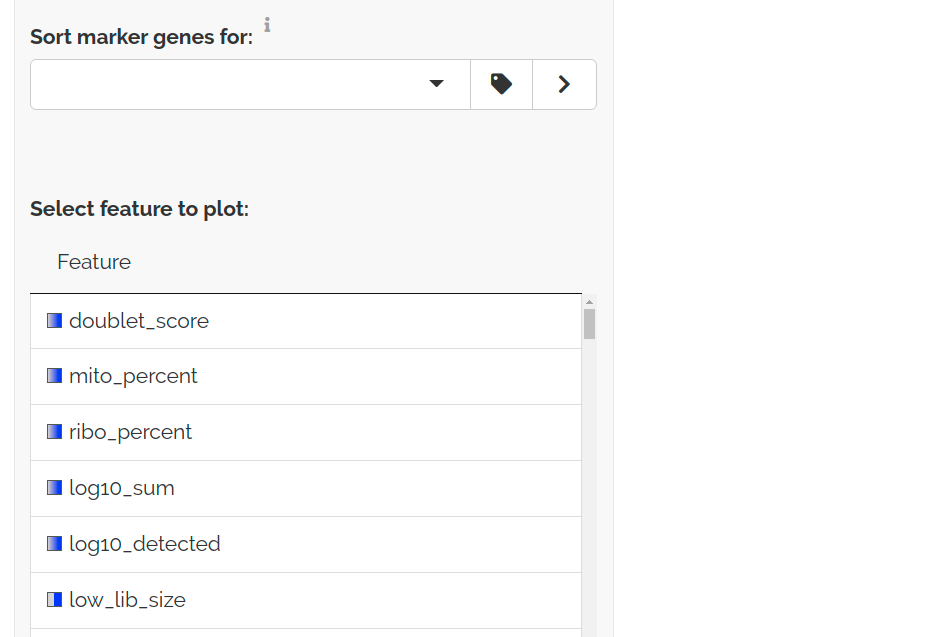
QC features are shown first when no cluster is selected
You can also plot and save new custom metrics. Saved boolean features can be used for further QC by subsetting.
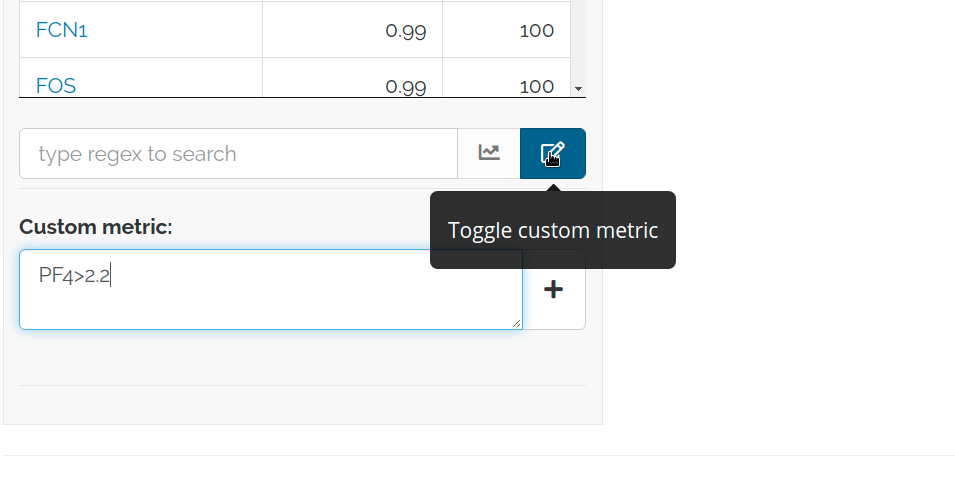
Custom metric examples:
- PF4>2.2: PF4 logcounts higher than 2.2
- PF4&IGF1: both PF4 and IGF1 are expressed
- PF4>2&IGF1>1: both have logcounts above specific threshold
- PF4>2.2&(cluster==1): PF4 logcounts higher than 2.2 in cluster 1
- PF4*IGF1>2: multiplication of PF4 and IGF1 logcounts are above 2
- PF4>2.2: PF4 logcounts higher than 2.2
- PF4&IGF1: both PF4 and IGF1 are expressed
- PF4>2&IGF1>1: both have logcounts above specific threshold
- PF4>2.2&(cluster==1): PF4 logcounts higher than 2.2 in cluster 1
- PF4*IGF1>2: multiplication of PF4 and IGF1 logcounts are above 2