Run Queries
How to run drug and genetic perturbation queries.
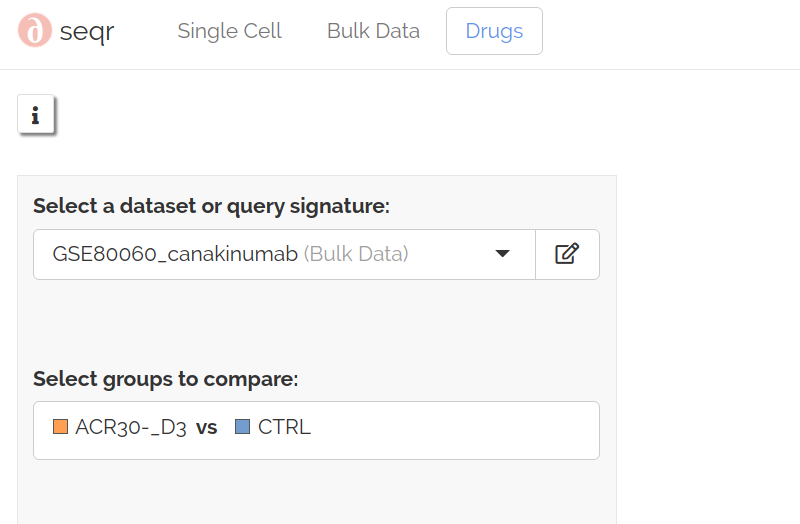
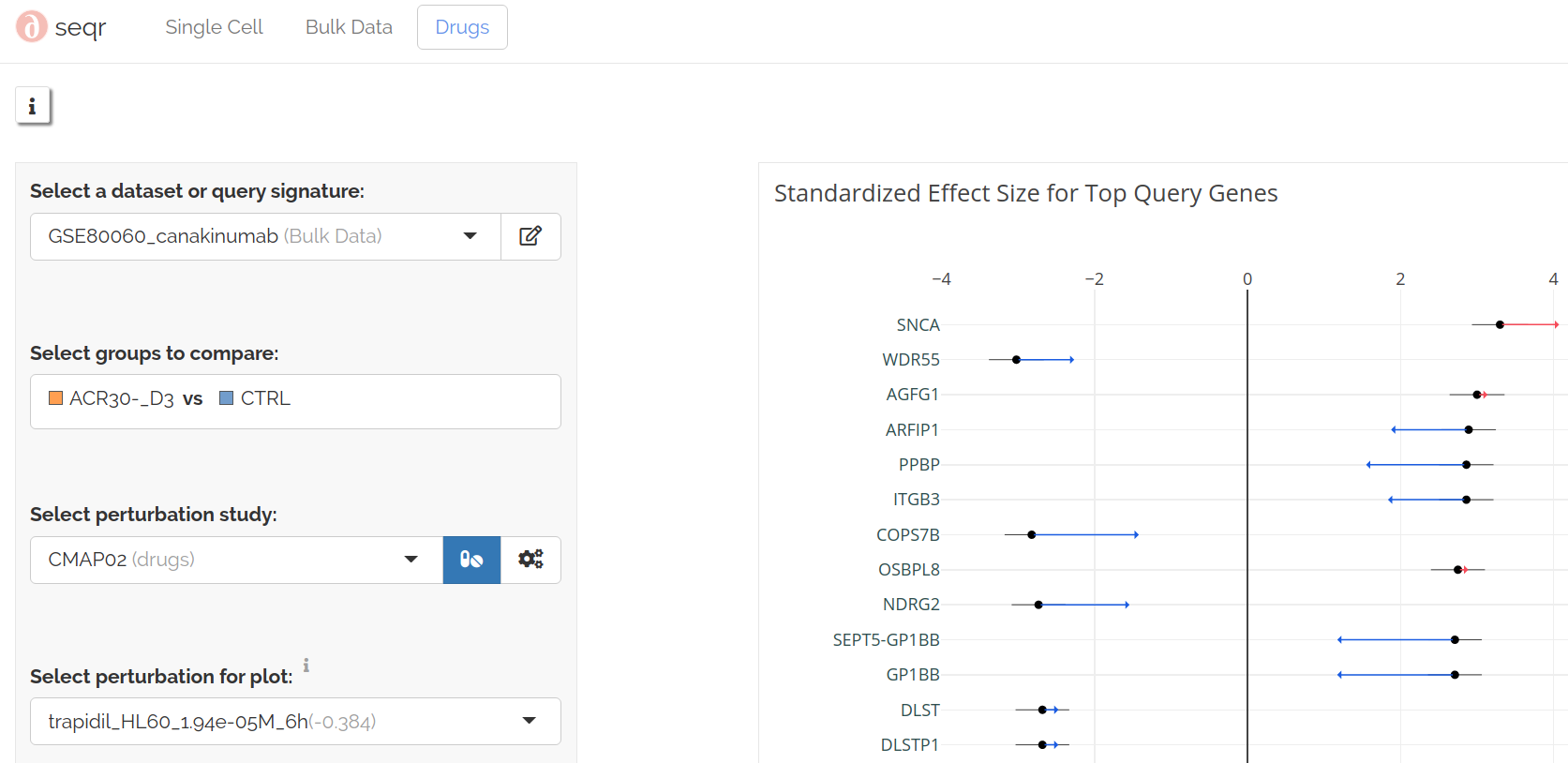
Select a query signature
To specify a query signature, select either an integrated single-cell dataset or a bulk dataset. If you select a single-cell dataset, select a cluster to use as your query signature. For bulk datasets, select a test and control group to form your query signature:
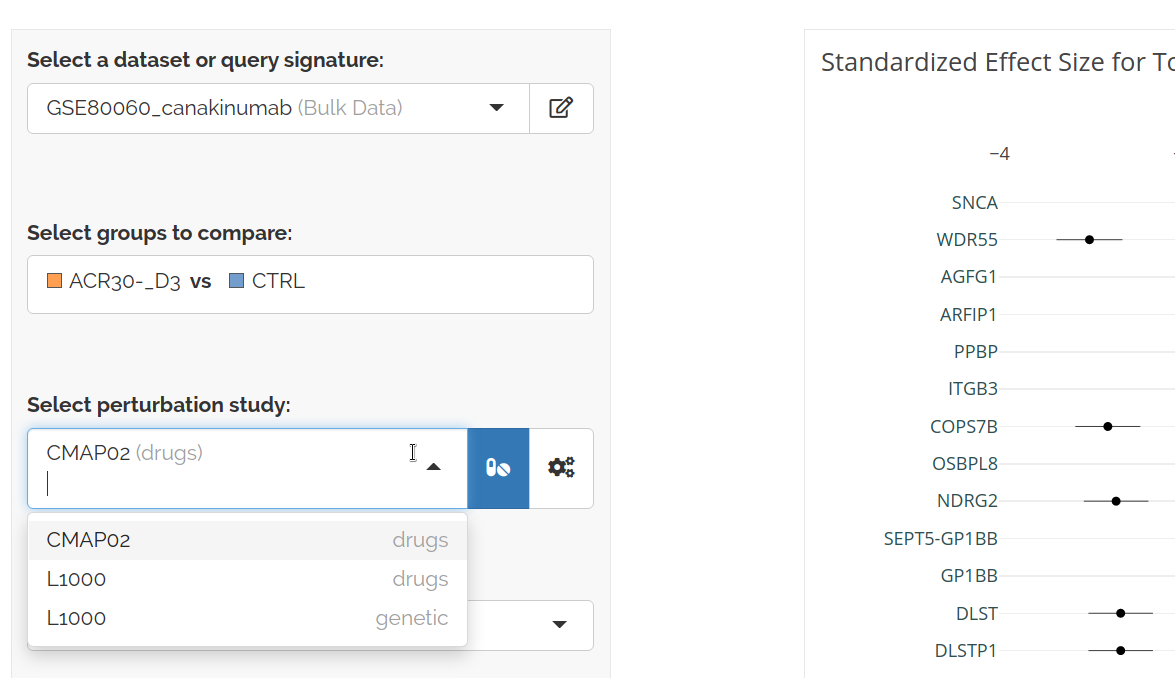
Select perturbation study
Next select a perturbation study to explore:
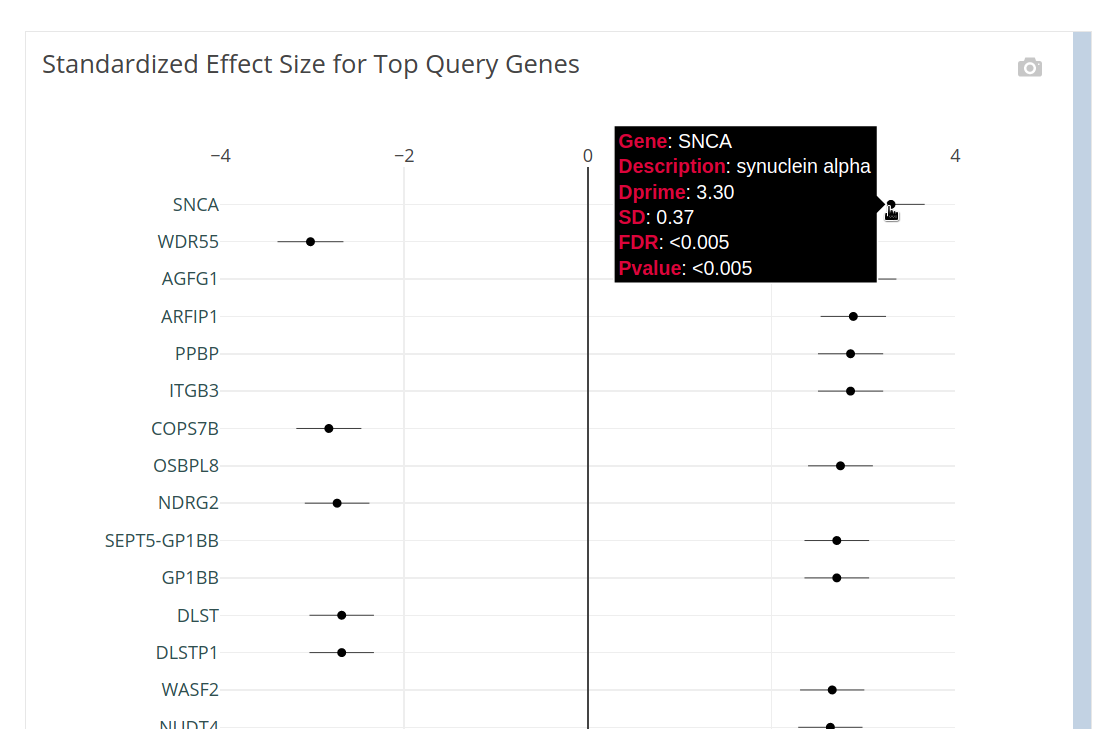
The differential expression of genes from your query signature are shown:
Explore results
A sorted table of top results is shown:
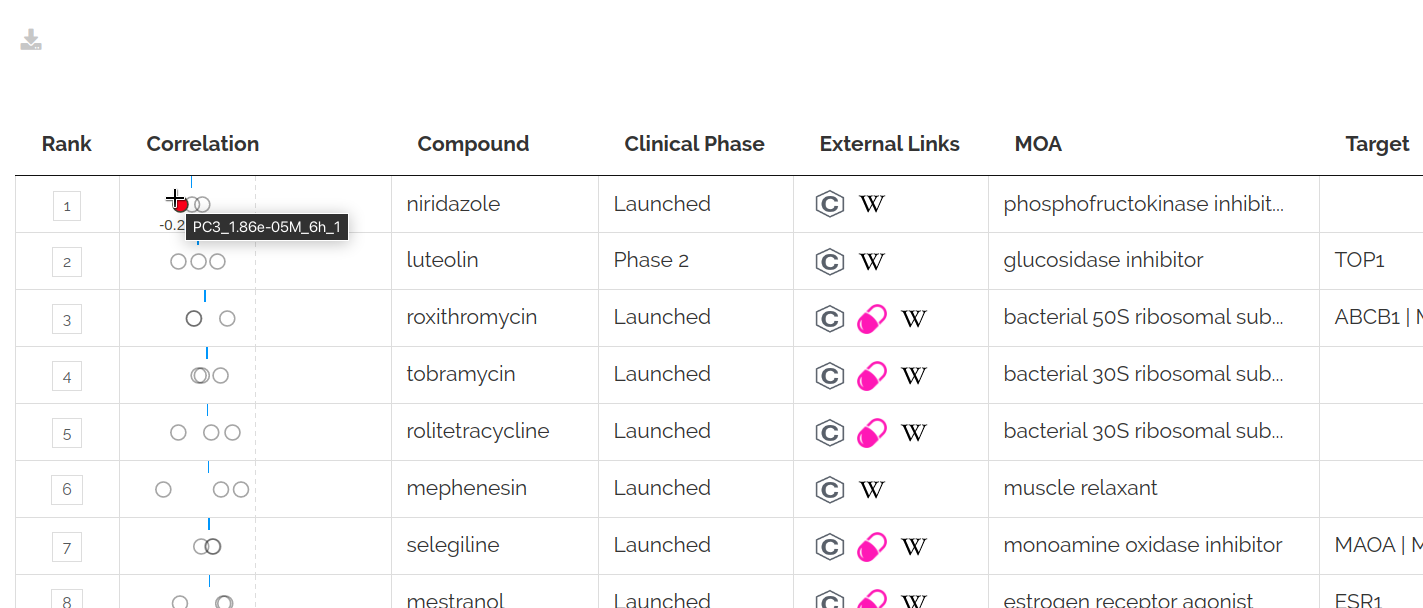
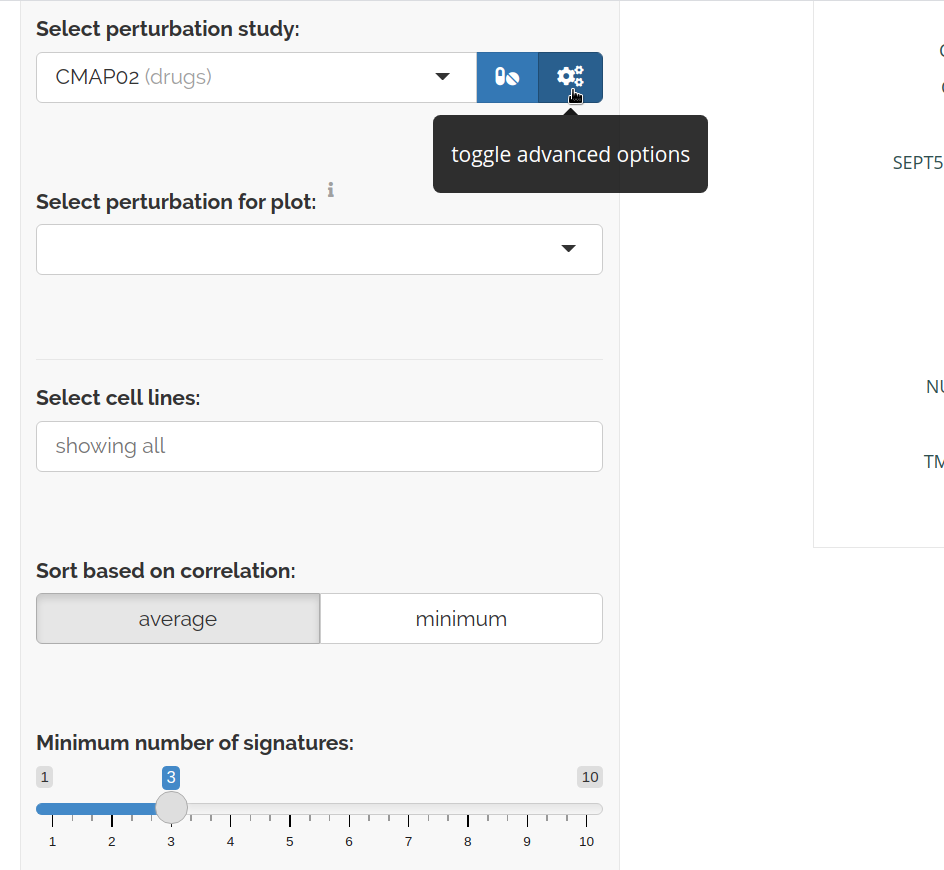
You can change the table sorting and filter for specific cell lines by clicking the advanced options toggle:
Plot perturbation effect
To view the predicted effect of a perturbation on the query genes, Select a perturbation for plot:
Perturbation as query
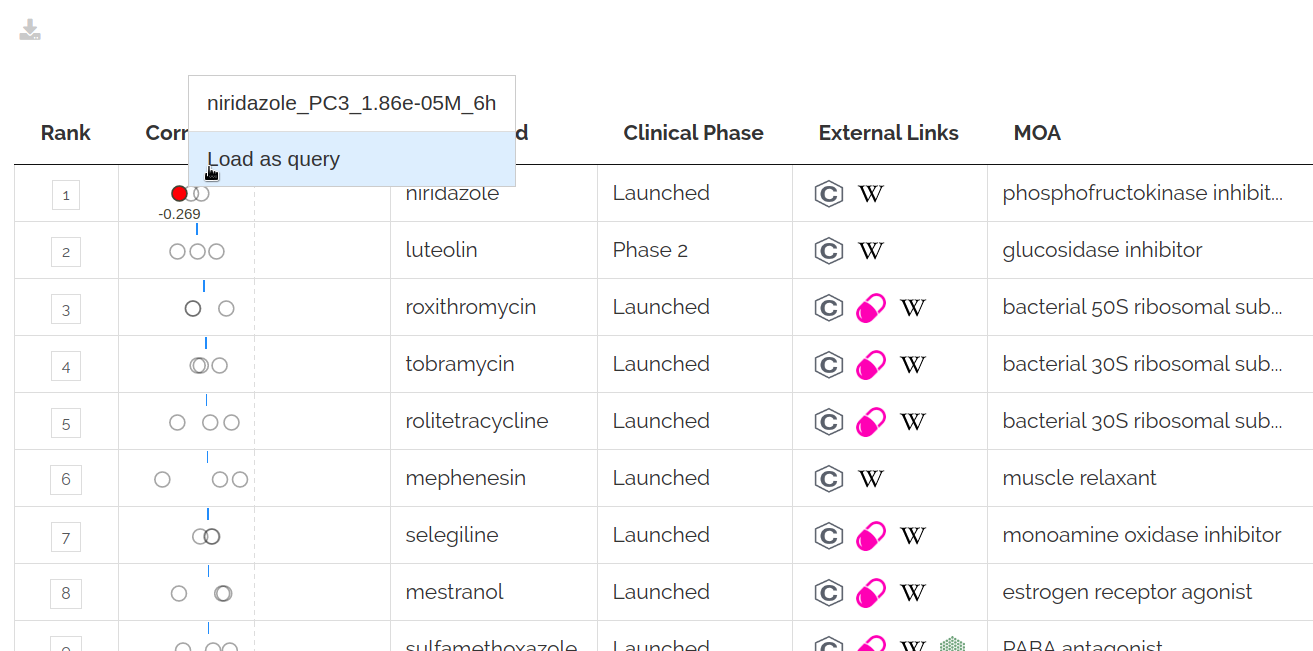
You can use expression profiles from the reference datasets to query for similar and opposing signatures in the reference datasets.
To do so either search for a signature in the Select a dataset or query signature input or right click on a correlation point and click Load as query: