Multi Sample Comparison
Compare expression, abundance, and pathways between test and control samples.
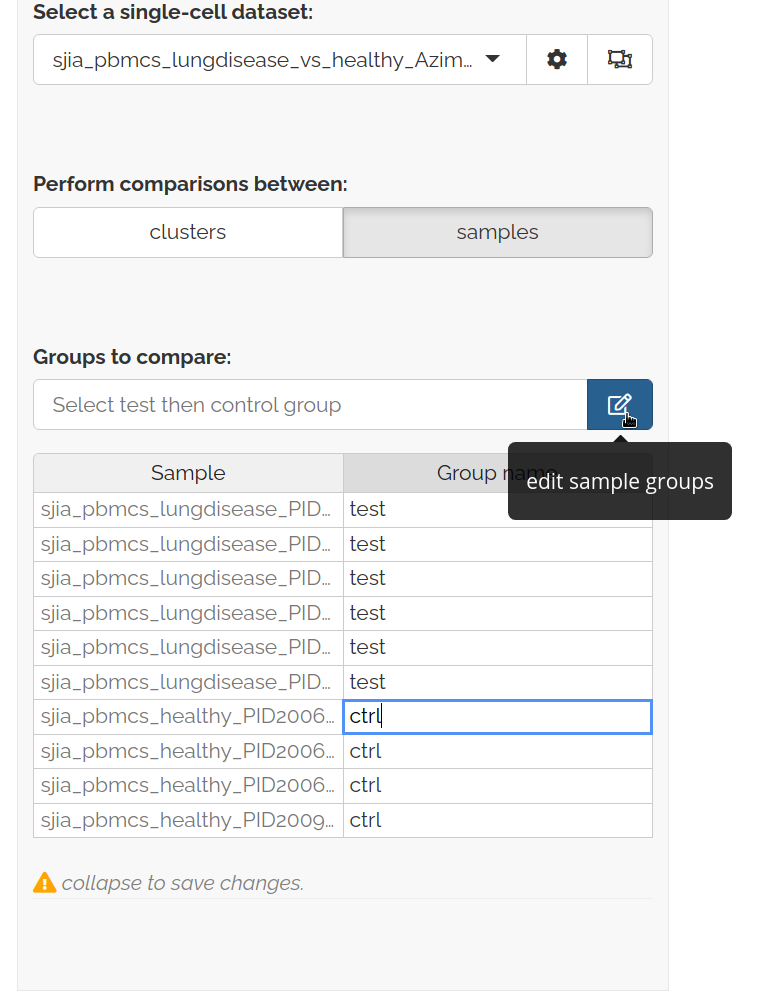
Select groups to compare
To run differential analyses between test and control samples:
- select an integrated dataset
- click samples comparison
- Open, fill out, and close the edit sample groups table
- Then specify test and control groups to compare
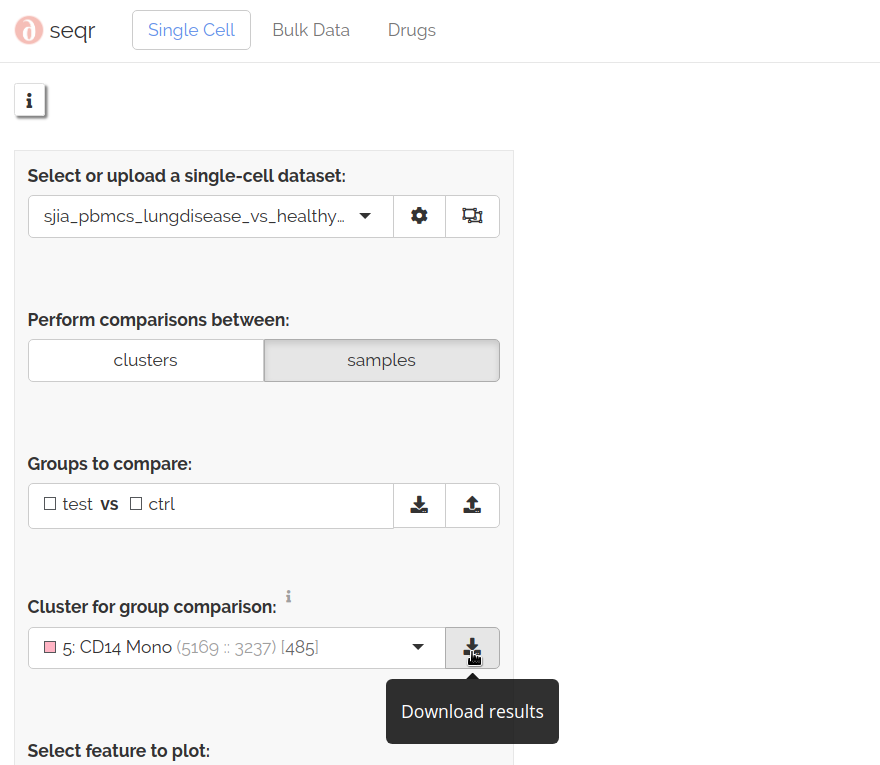
Download differential analyses
To download the results of differential analyses between test and control samples select a cluster for group comparison then click download:
Pseudobulk differential expression
Pseudobulk expression profiles are used for differential expression analyses between test and control groups.
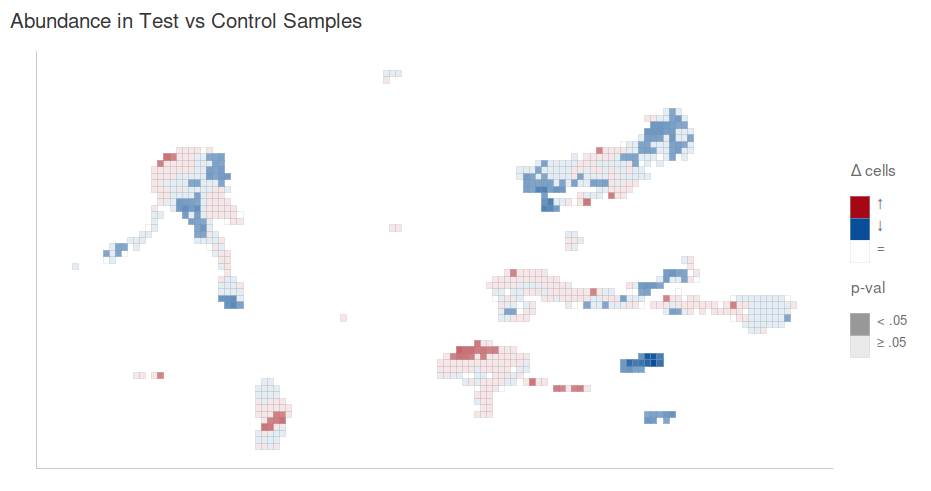
Differential abundance
A differential abundance analysis is included in the downloaded results for any cluster. A grid-based differential abundance plot also allows you to visualize regions of differential abundance:

Pathway analyses
Dseqr runs Gene Ontology over-representation analysis of significantly up- and down-regulated genes. This uses a cached adaptation of goana.
- Uses genes with FDR < 0.05 with a min of 50 up/down.
- Uses expressed genes as a background (from edgeR::filterByExpr).
- Removes terms with fewer than 4 up/down genes (too few).
- Removes terms with more than 250 genes (too broad).
- Removes terms with Δup-down < 3 genes per 10 (not distinctly up/down).
- Removes terms with FDR > 0.05 (not significant).
- Removes terms with FDR < 0.05 for both up/down (not distinctly up/down)
Visual exploration
With samples comparison, select a gene to see expression side-by-side in test and control cells:
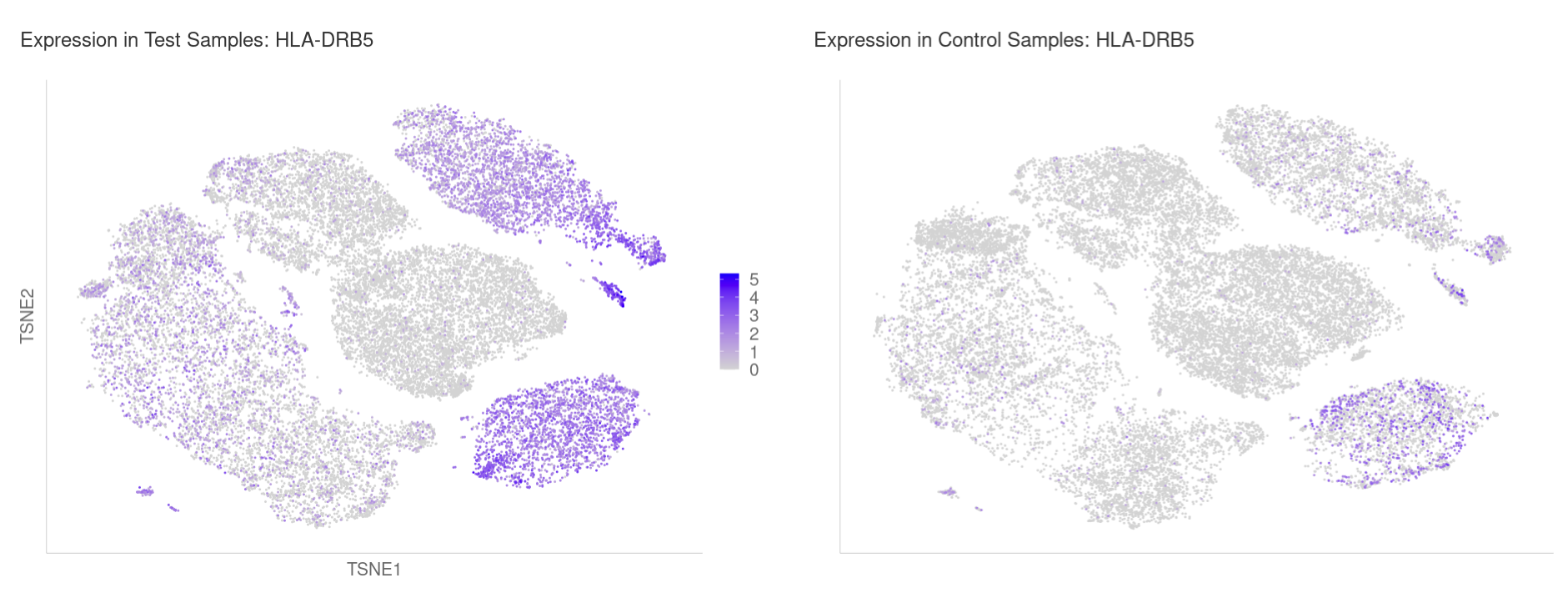
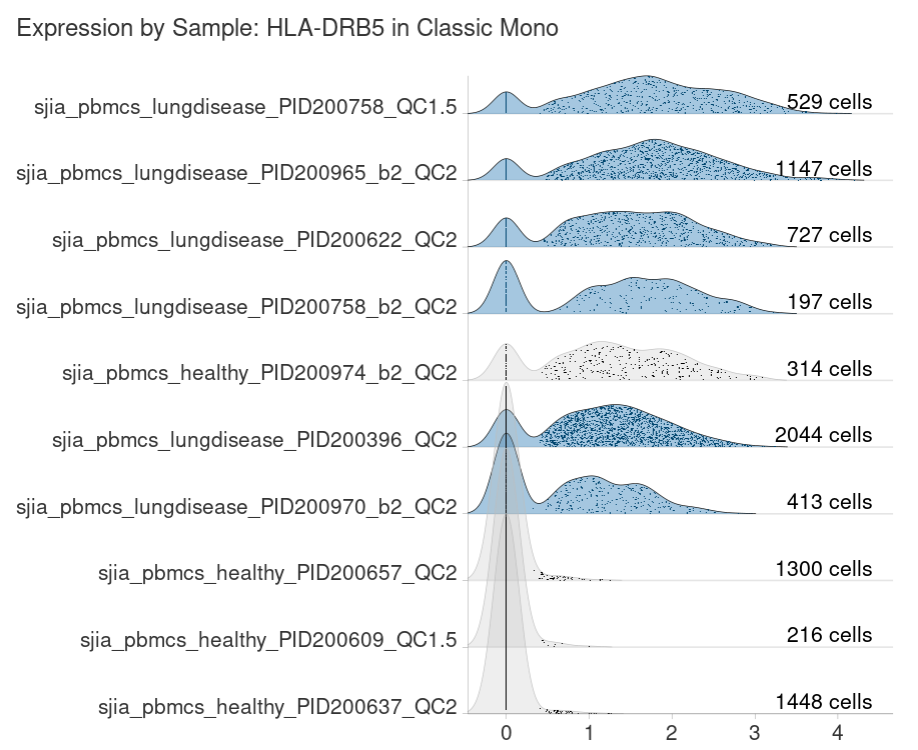
You are also shown the distribution of logcounts across each sample for the selected cluster:
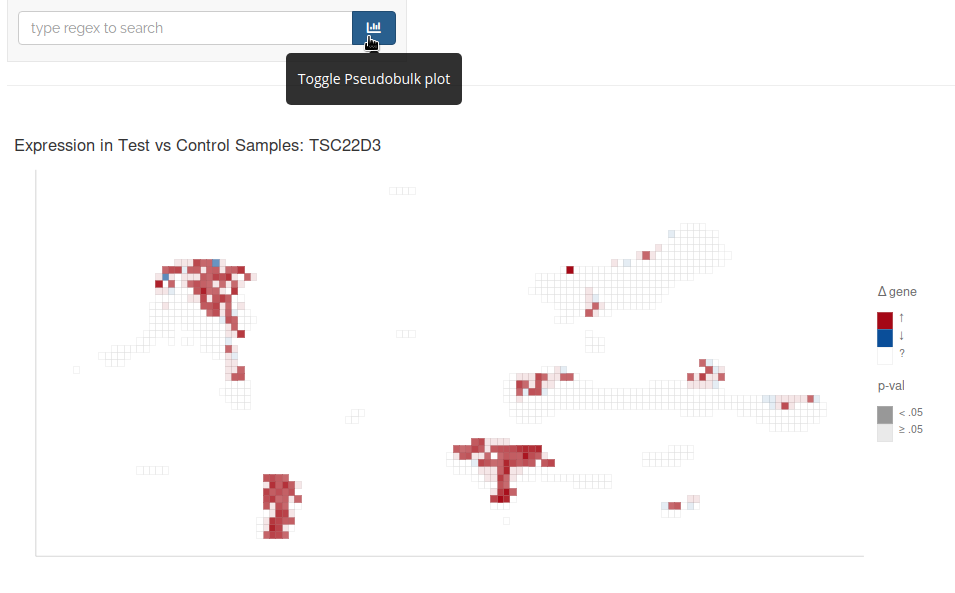
You can also toggle the pseudobulk plot button to visualize regions of differential expression for the selected gene:
Compare all clusters
To view genes that are consistently differentially expressed across clusters select All Clusters:
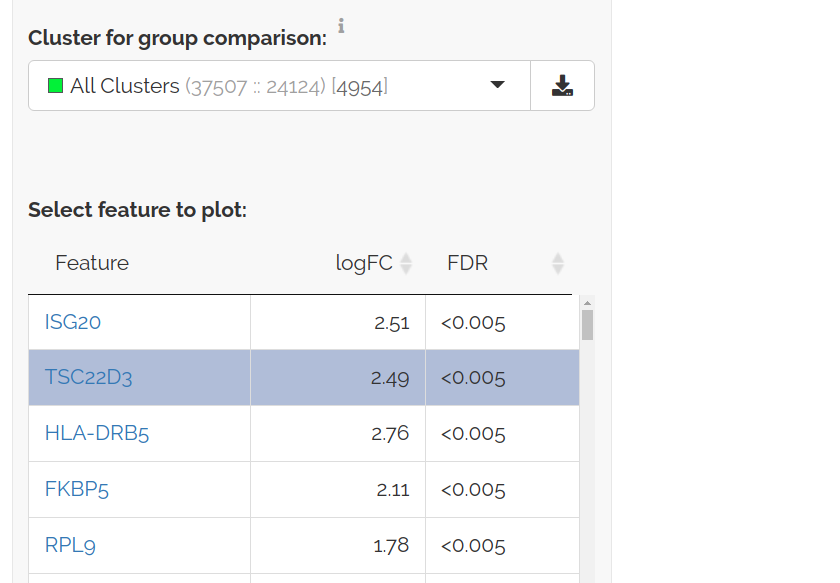
FDRs are similarly calculated with a p-value based meta-analysis using Stouffer's method followed by adjustment for multiple hypothesis testing.